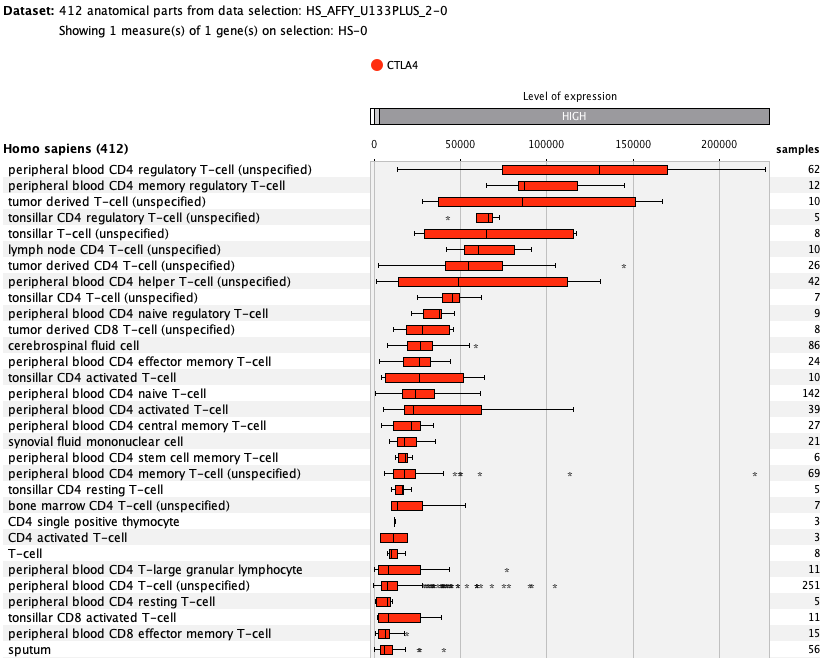
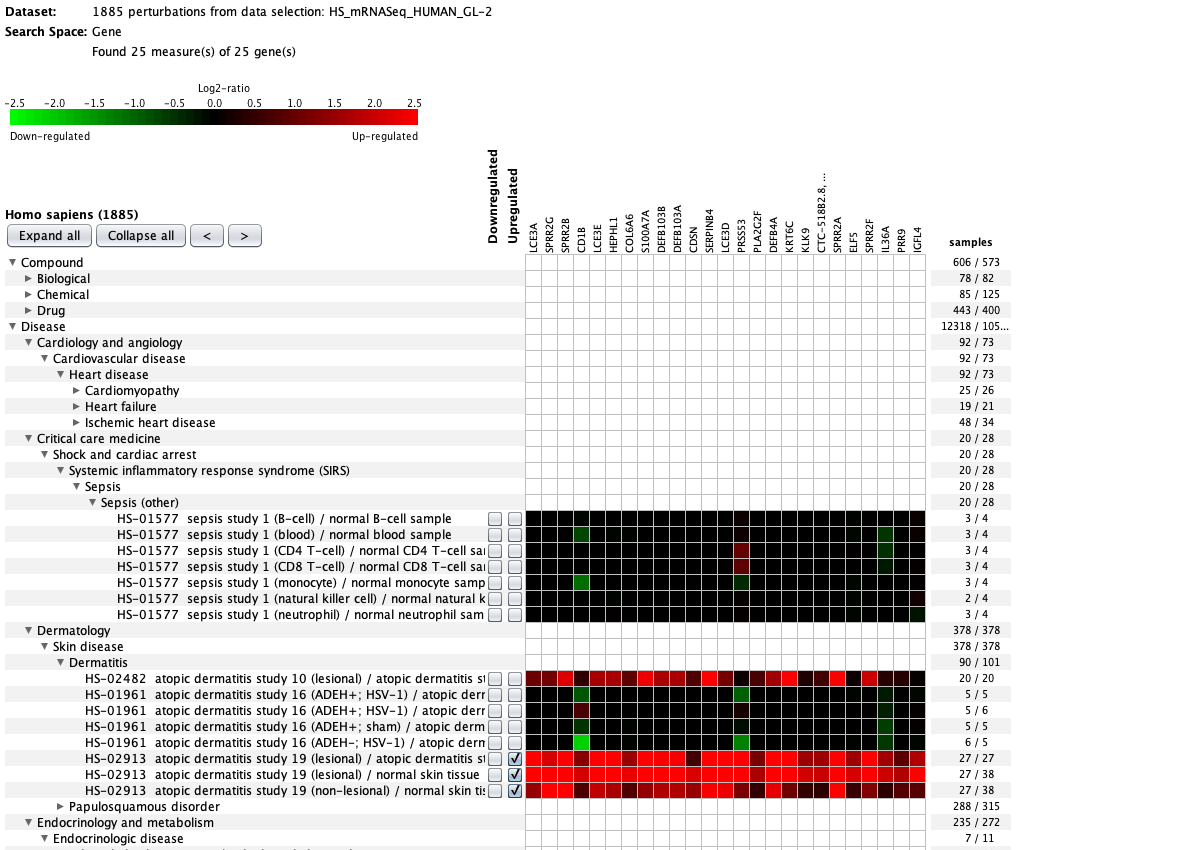
We curate public and private studies
from bulk tissue to single cells
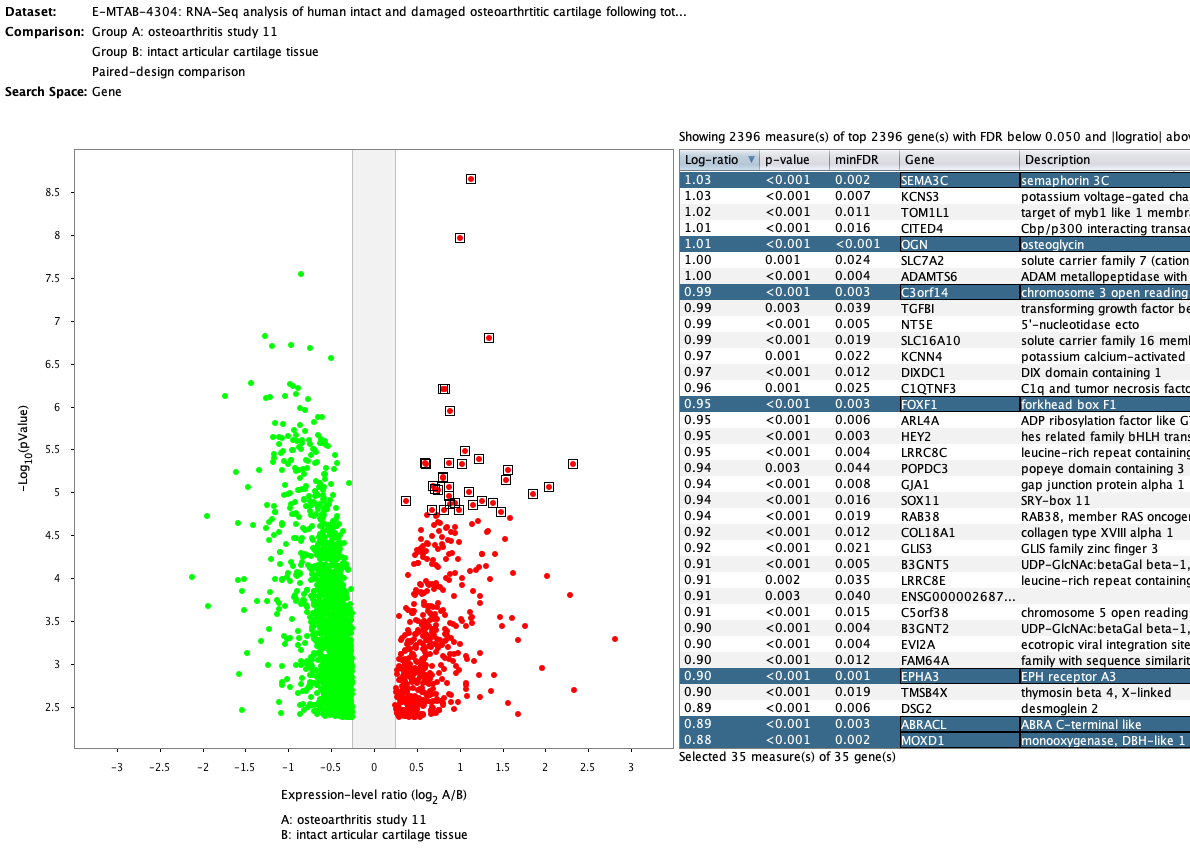
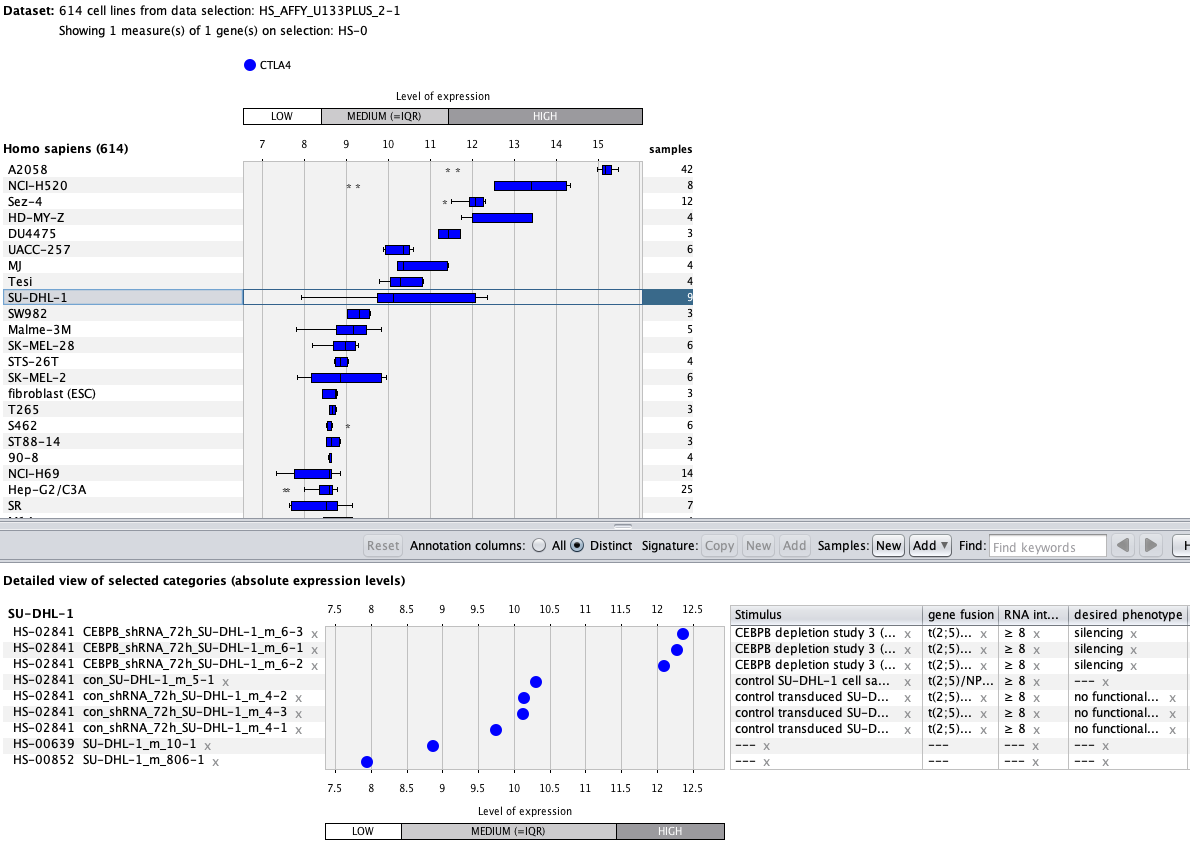
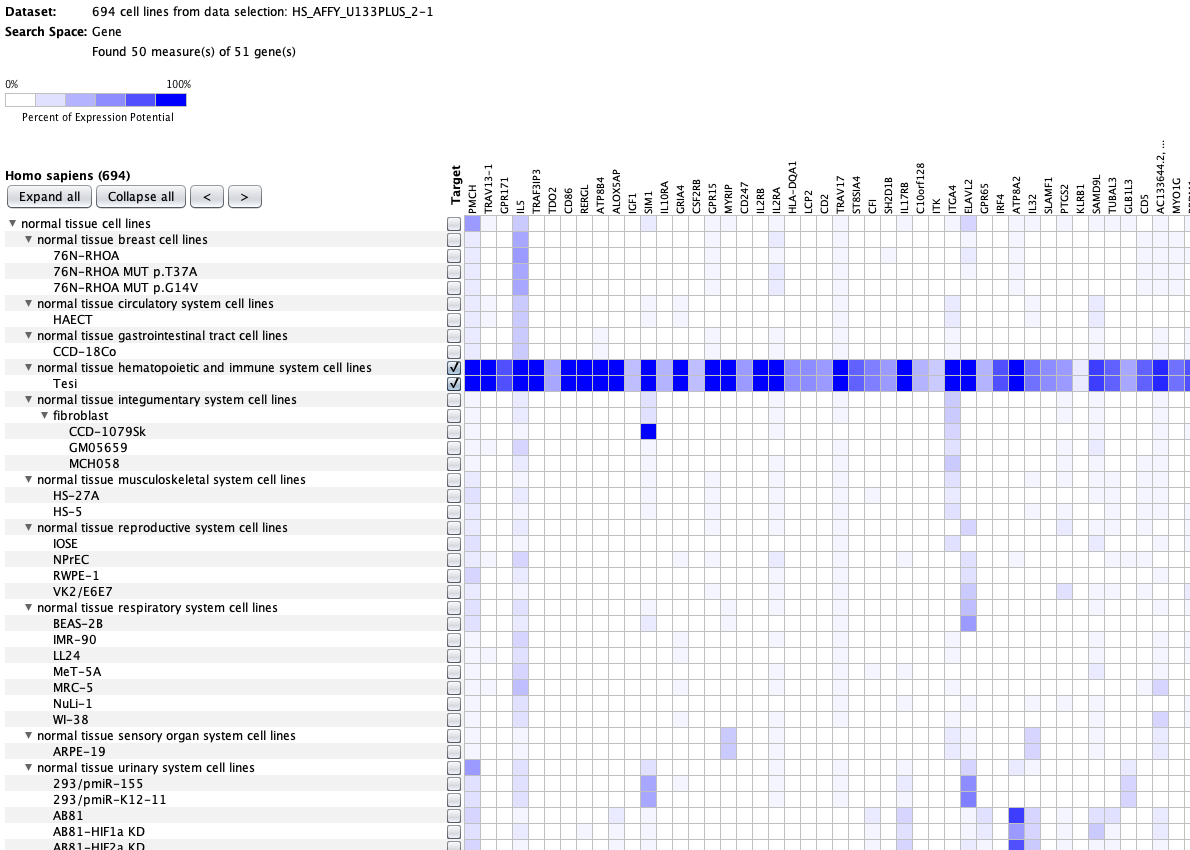
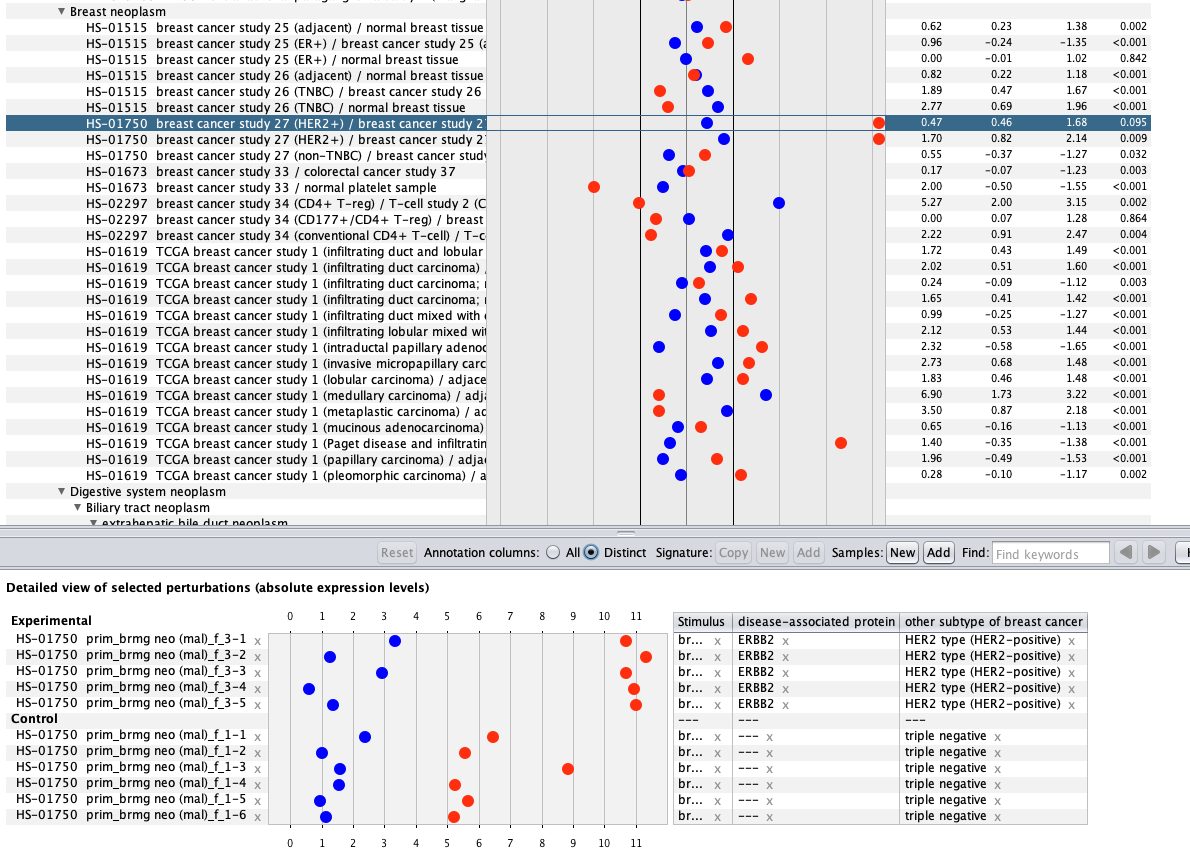
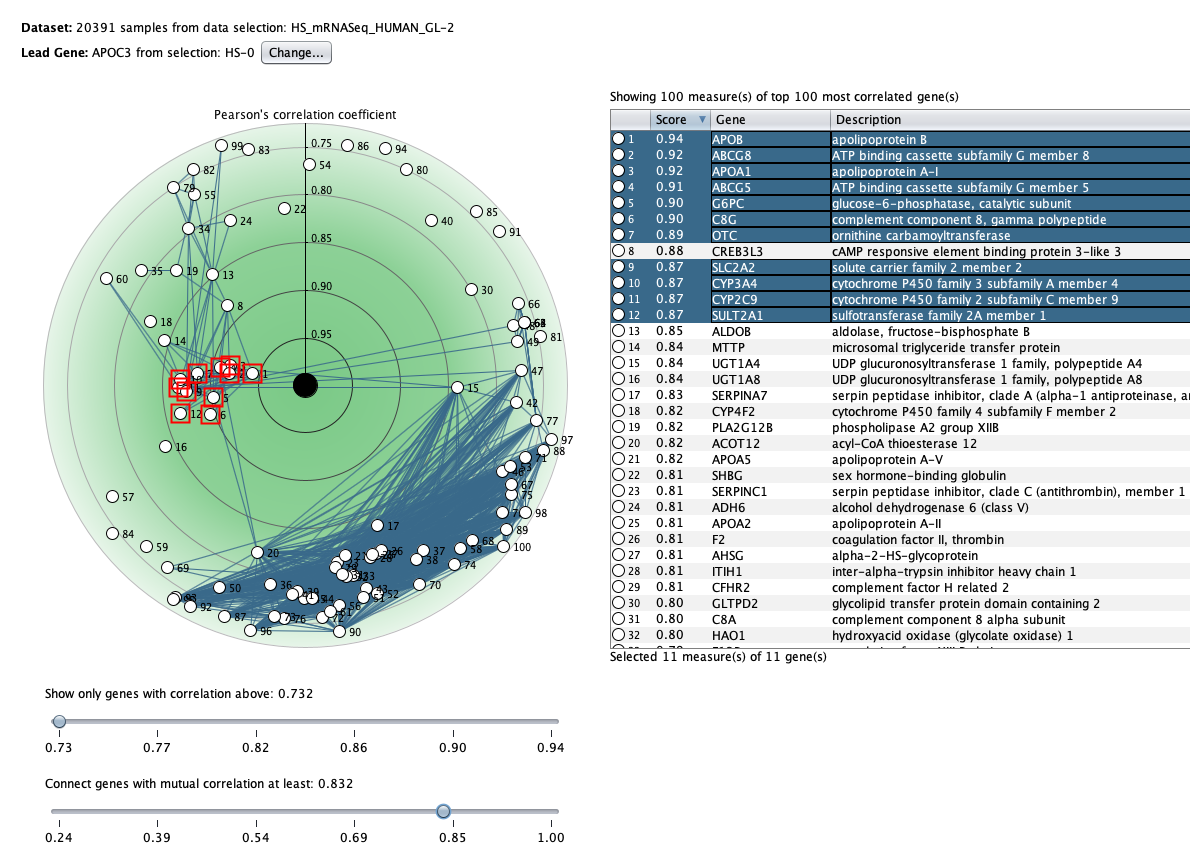
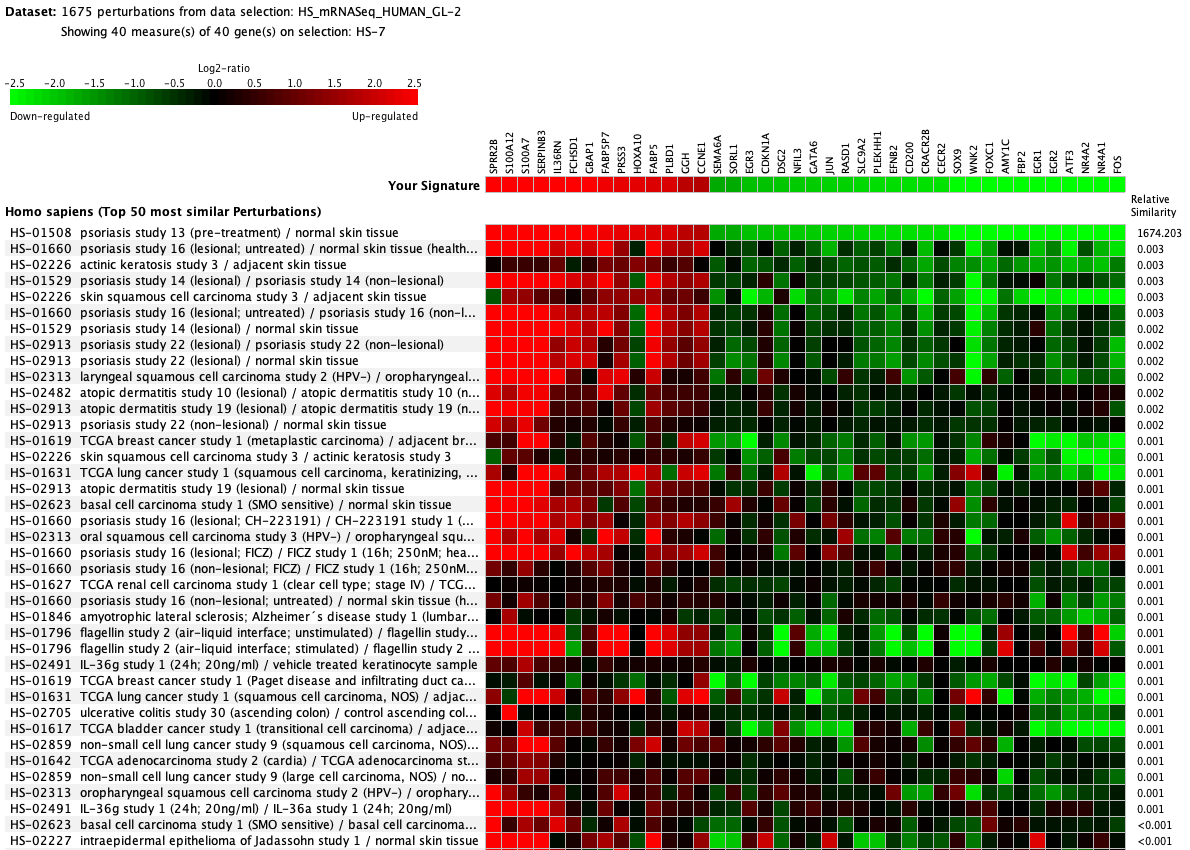
Over more than a decade, we have continuously perfectioned techniques and vocabularies for the biocuration of biological experiments. Our unique collections of deeply curated and high quality transcriptomic data, combined with our high-performance but easy-to-use tools, enable global and instant analysis of the world's transcriptomic data for novel discoveries and for validation.