The below example study illustrates an analysis done with GENEVESTIGATOR® with a 10x RNA-Seq dataset that has been curated by the teams at NEBION. The curation included data processing, quality control, (sub)clustering, cell-type identification and cross-study harmonization, metadata annotation, and integration into the platform for visualization. A particular feature of the 10x data is the aggregation of cells with identical annotation (cell type, cell state, subject ID and conditions). 10x data can be visualized on the aggregate level in GENEVESTIGATOR®.
Study details
- GEO accession: GSE145926
- Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020 Jun;26(6):842-844
- 10x, aggregate level
- Healthy/moderate/severe COVID-19 patients – cells from broncho-alveolar lavage fluid (BALF)
- 58534 high-quality cells
Cell type analysis
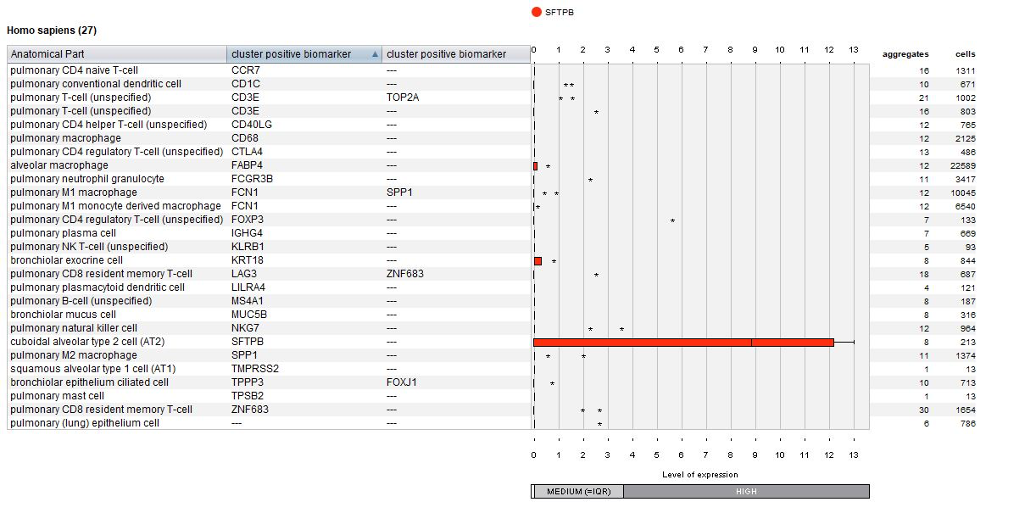
The below plot displays the summary view from the GENEVESTIGATOR® Samples tool, as generated using the “Plot groups…” feature or selecting the corresponding metadata column in the list of samples. As an example, we chose to visualize the expression of gene SFTPB across all cell types identified in this study. SFTPB shows specific expression in cuboidal alveolar type 2 (AT2) cells.
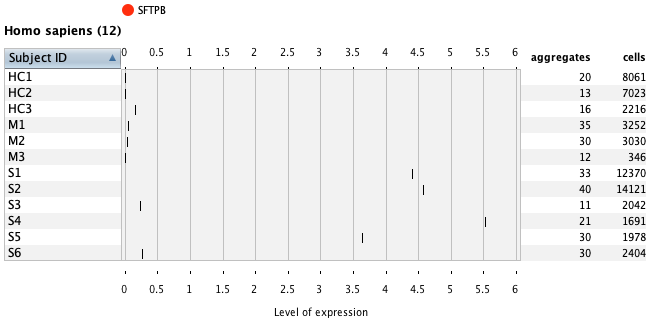
Using the Samples summary view again, one can easily look at gene expression in any chosen cell type(s) under different conditions or patients. In the below example, as a general overview one can check the number of cells measured for each subject across all measured cell types.
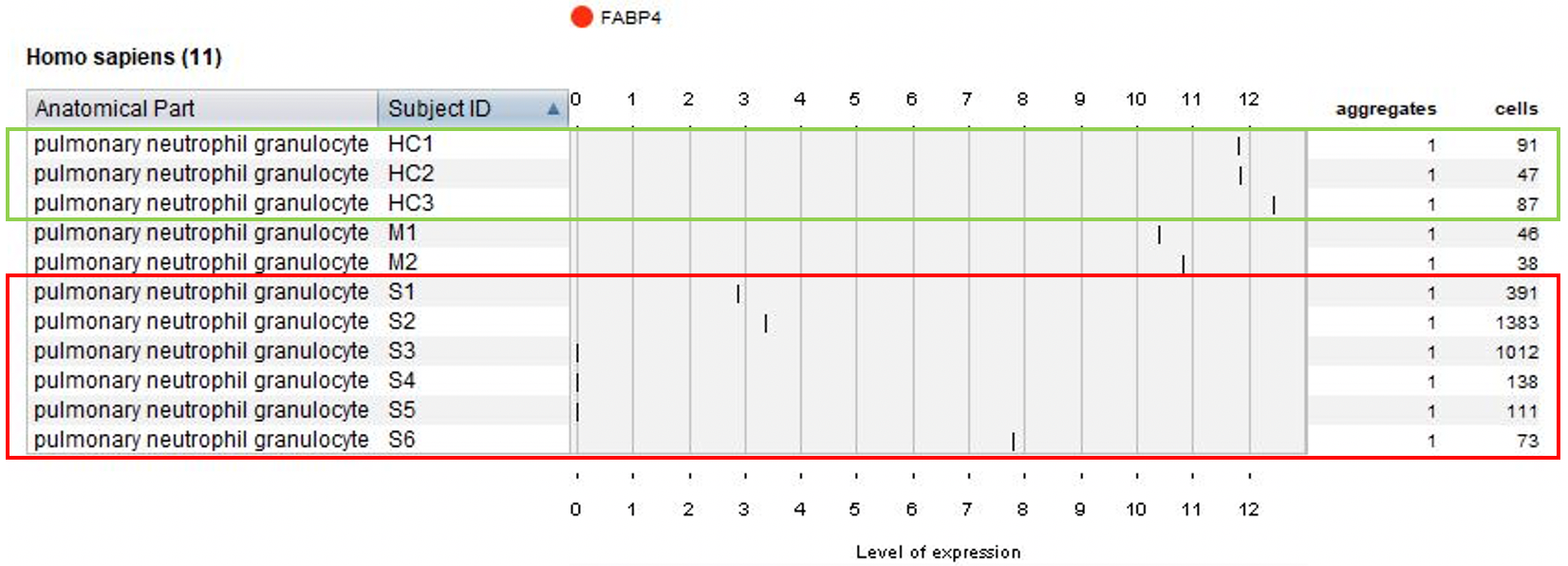
Next, we visualized the expression of FABP4 and the number of pulmonary neutrophil granulocyte cells across all patients.
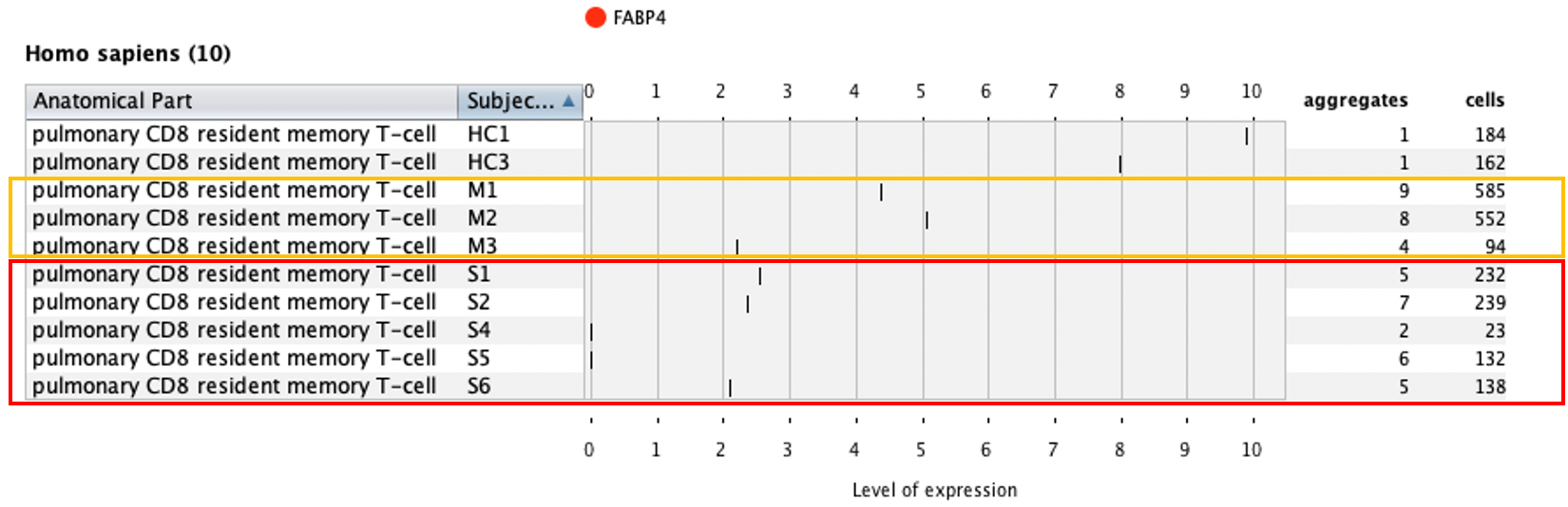
Additionally, we showed that CD8 resident memory cells are increased in moderate cases but not nearly as much in severe cases – these are two important findings of the original publication.
Differential expression analysis
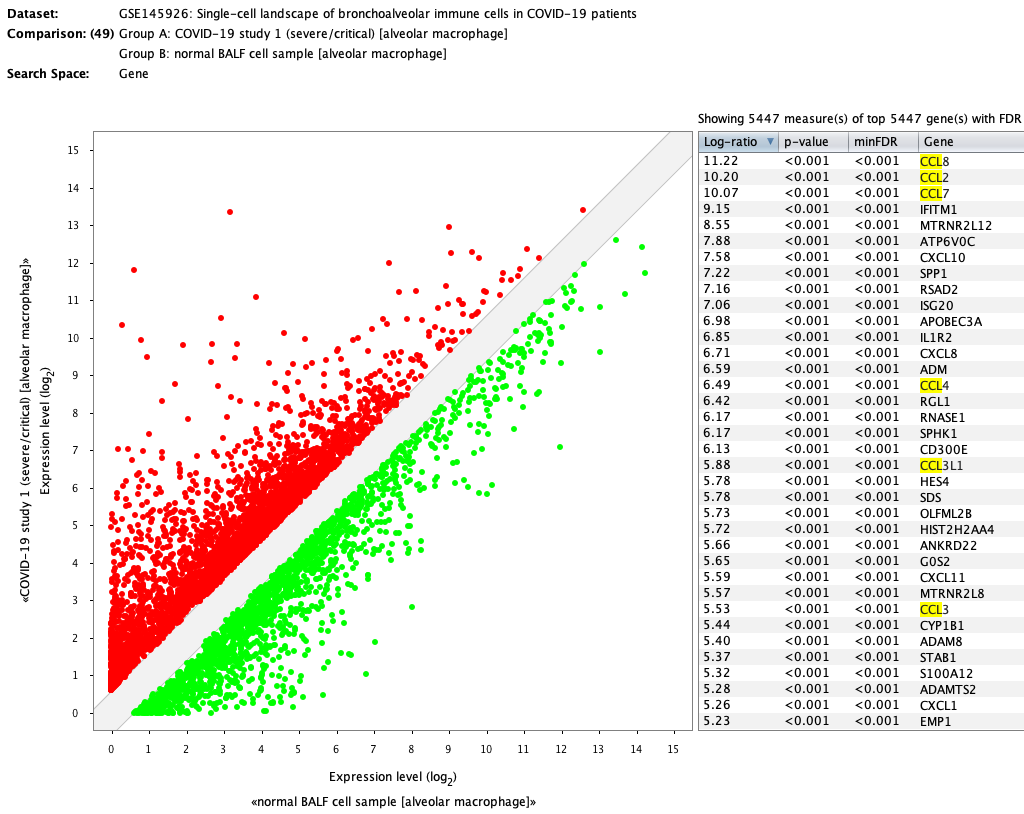
The differential expression carried out in the publication can be repeated in GENEVESTIGATOR® within seconds by simply choosing the corresponding comparison (severe alveolar macrophages (6 aggregates) against healthy alveolar macrophages (3 aggregates)). The results are very similar to the ones obtained in the original publication. Genes from the CCL family are highlighted.
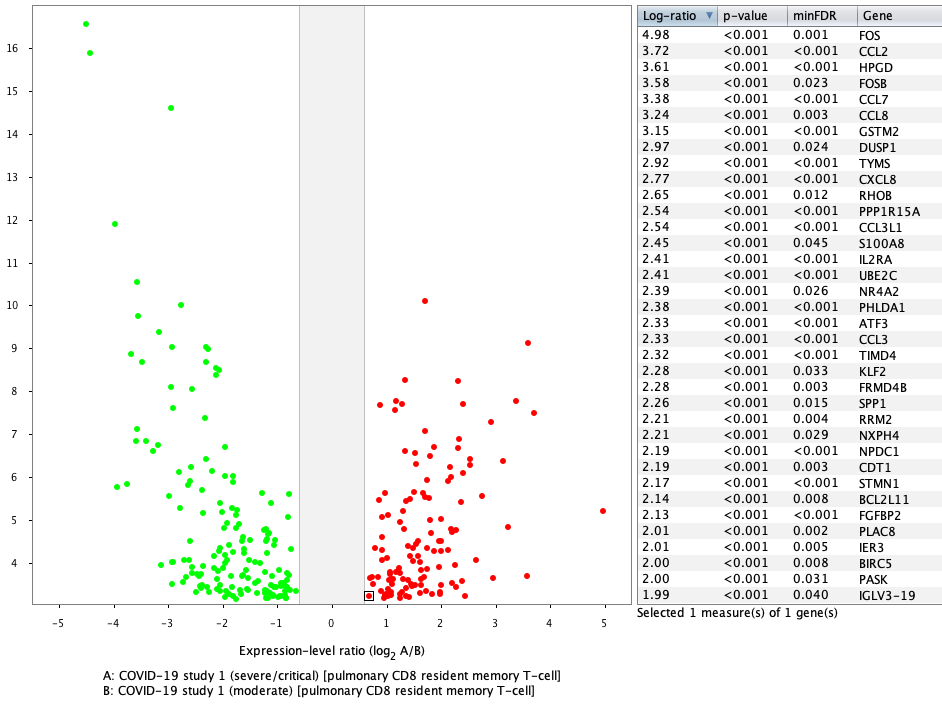
Differential expression of aggregated CD8 T cells (pulmonary CD8 resident memory T-cell) between severe and moderate cases in GENEVESTIGATOR® yields very similar results to the original publication, where MAST, an algorithm designed for single-cell differential expression analyses, was used. In this case, the results are displayed as a volcano plot.
Validation of candidate genes
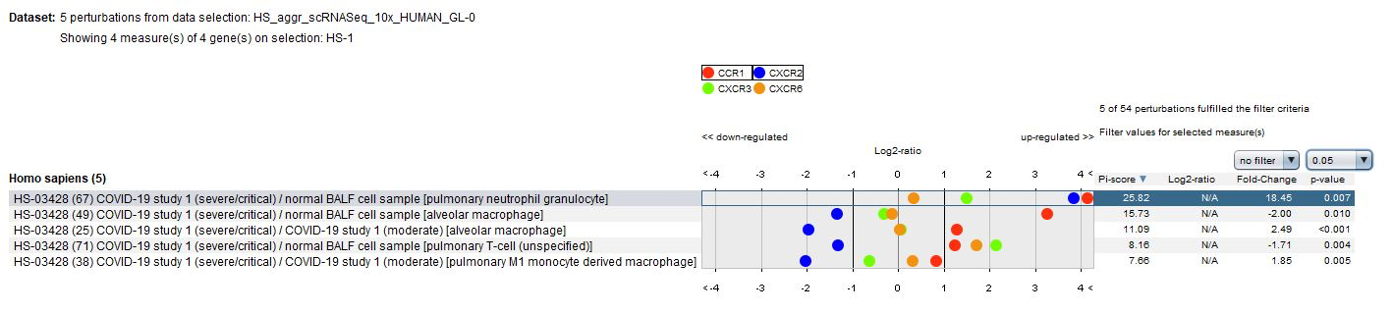
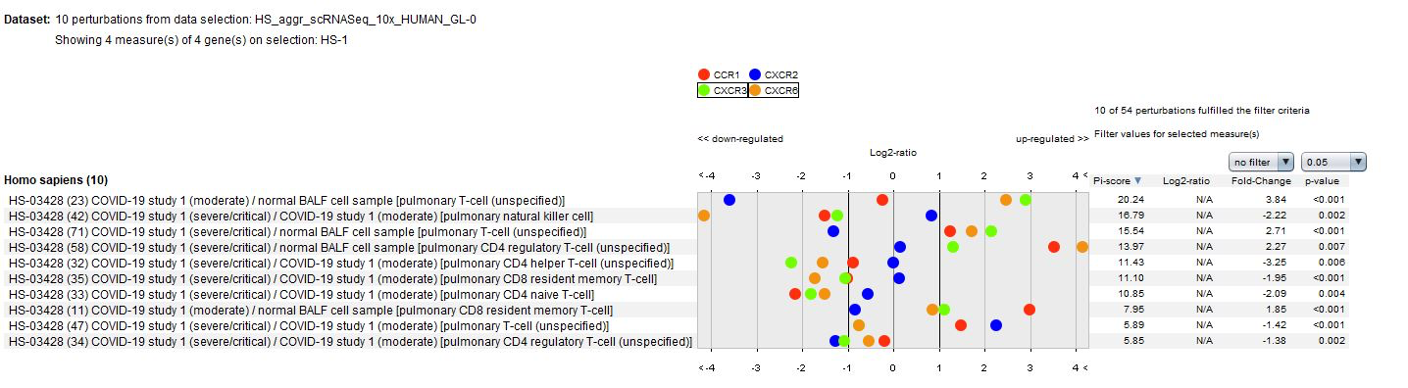
Data from the publication suggest that lung macrophages in patients with severe COVID-19 infection may contribute to local inflammation by recruiting inflammatory monocytic cells and neutrophils through CCR1 and CXCR2, whereas macrophages in patients with moderate COVID-19 infection produce more T cell-attracting chemokines by engaging CXCR3 and CXCR6. These results could be easily confirmed using the GENEVESTIGATOR® Perturbations tool in CONDITION SEARCH tools.
Summary
With GENEVESTIGATOR®, single-cell scientists and biologists without specific single-cell training can easily visualize and analyze single-cell studies from the 10x platform on the aggregate level. In this example, we were able to:
- Easily analyze 58534 cells isolated from healthy/moderate/severe cases of Covid-19 patients (all tools)
- Perform fast and clear visualization of distribution of immune cells across different patients (Samples tool)
- Easily and reliably run differential expression calculations in any cell types/conditions (Differential Expression tool)
- Rapidly identify cell types/conditions with significantly changed expression of any gene of interest (CONDITION SEARCH Perturbations tool)
- Obtain information about V(D)J annotations (tooltips)
Access to curated single-cell studies
The presented study is freely available on GENEVESTIGATOR® for all users for demonstration purposes. Various compendia of deeply curated single-cell studies from the Smart-Seq or 10x platforms are available upon subscription. To access our deeply curated compendia on GENEVESTIGATOR®, please contact us at sales@nebion.com.