SARS-Cov-2 differs from closely related coronaviruses (including SARS-Cov) in the sequence of its spike protein, among other differences. Unlike SARS-Cov, the newly emerged coronavirus SARS-Cov-2 spike protein contains a multibasic furin-like S1/S2 cleavage site. A recent study has identified that cleavage of the spike protein at this site is required for SARS-CoV-2 virus entry into human lung cells (Hoffmann et al., 2020).
Variation around the envelope protein cleavage site plays an important role in tissue tropism and pathogenicity of several viruses (Braun and Sauter, 2019). For instance, highly pathogenic influenza A viruses harbour a multibasic furin cleavage site, while their low pathogenic counterparts do not. In GENEVESTIGATOR®, one can easily explore the tissue expression profile of the priming protease furin (PCSK3), or other furin-like proteases. Using the 2-gene plot tool, we have shown a rather uniform expression of furin across tissues and cell types (Figure 5). Interestingly, this broad expression profile is in line with the observed broad tissue tropism of the newly emerged virus, as detected in COVID-19 patient autopsy tissue samples (Puelles et al., 2020).
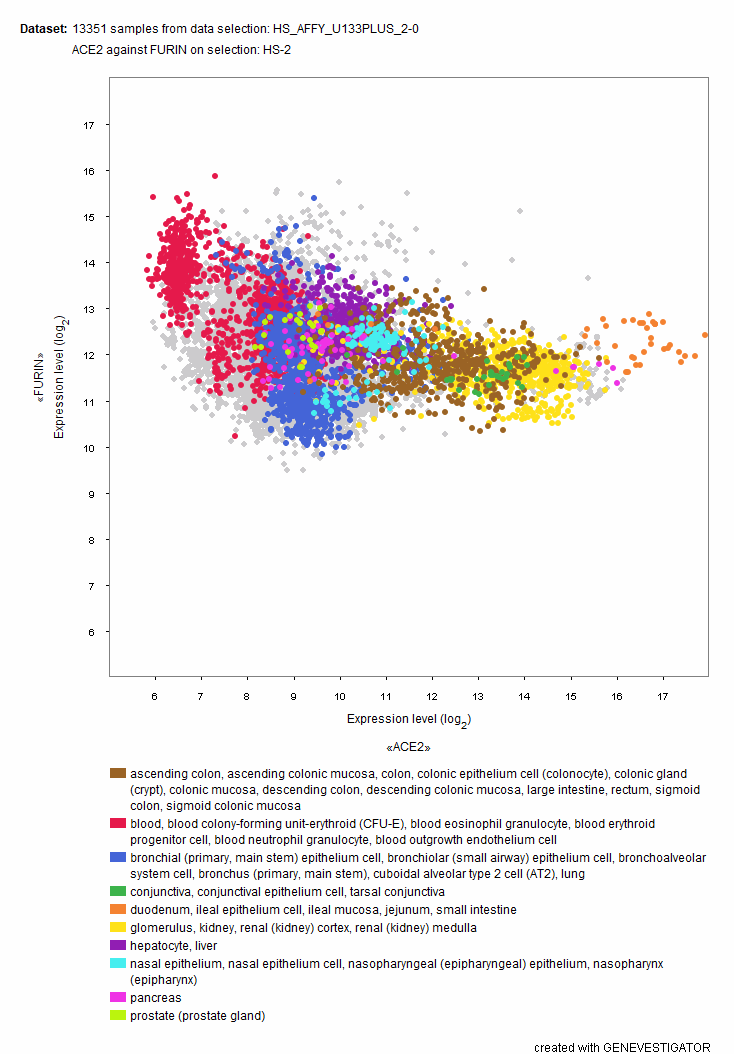
Figure: GENEVESTIGATOR® 2-Gene scatterplot showing the expression levels of the priming protease furin plotted against the receptor ACE2 in 13351 samples from healthy tissue and primary cells (Affymetrix Human 133 Plus2 compendium). The priming protease furin shows a ubiquitous expression in all tissue and cell types examined.
As mentioned above, the enzymatic activity of furin is known to be exploited by numerous viral and bacterial pathogens, including HIV and highly pathogenic strains of influenza. Several proteins expressed by infected cells have been shown to suppress this furin-mediated processing of viral proteins (Braun and Sauter, 2019). We have explored the regulation of three of them, GBP2, GBP5 and PAR1/F2R in response to drug/compound treatment. Among the top-20 compounds up-regulating GBP2 and GBP5 proteins, we observed several Toll-like receptor agonists such as poly I:C or LPS. Surprisingly, we also found that decitabine, a nucleic acid synthesis inhibitor, up-regulates expression of all the three antiviral proteins studied.
It is tempting to speculate that dependency on host priming proteases is one of the key determinants of tissue tropism and spread of the newly emerged SARS-Cov-2 virus. They may thus represent potential targets for therapeutic intervention. Their expression and regulation certainly warrant further investigation.
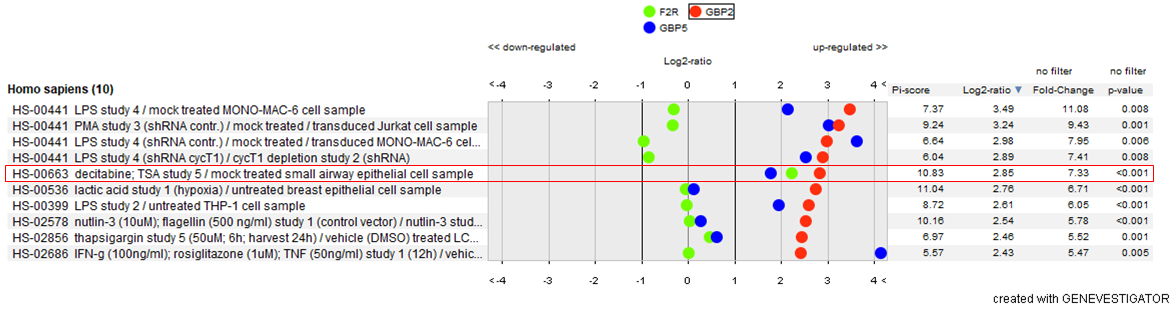
Figure: Top-10 compounds up-regulating the expression of guanylate‐binding protein 2 (GBP2), a protein known to inhibit furin‐mediated viral protein processing. Effects of compound treatment was investigated in 6171 samples of tissues and primary cells (Affymetrix Human 133 Plus2 compendium).