A key single-cell RNA-Seq study for exploring the adult nervous system was published by Zeisel and colleagues. It is entitled Molecular Architecture of the Mouse Nervous System. In this study, cells from different regions of the adult murine central and peripheral nervous system were sequenced using a 10x Genomics Chromium protocol. The authors provide the data, as analyzed in the original publication, in an interactive web interface. The raw data is publicly available in the Sequence Read Archive as SRP135960.
In GENEVESTIGATOR®, this study is available on an aggregate level. Users who have licensed the single-cell compendium “Nervous System” will find it as MM-01239. Like all other studies in this unique compendium, this study was curated and integrated starting from the publicly available raw data. NEBION’s state-of-the-art processing and analysis pipeline uses the latest algorithms for cell clustering, batch-effect removal, and normalization. This resulted in a refined clustering and marker gene calculation, and ultimately in a more precise cell-type identification.
As part of NEBION’s curation, the metadata of this study were enriched using controlled vocabulary, including cell-level annotations. This consistency ensures that the present study can be analyzed on its own in GENEVESTIGATOR®, or as part of a compendium together with numerous other studies.
What molecular insights can be gained by analyzing this study in GENEVESTIGATOR®?
Analysis of public transcriptomic data in GENEVESTIGATOR® allows the user to investigate genes or conditions of interest at the highest level of resolution. At the same time, users can analyze large and complex single-cell studies within a few seconds and using a few clicks. All the valued tools for bulk-tissue data analysis are fully compatible with the single-cell data. Hence, there are many different workflows to investigate a single-cell study. For an example of such an analysis, see here.
In the following, we list some ideas for the analysis of this adult nervous system single-cell RNA-Seq study in GENEVESTIGATOR®:
- Find cell-type-specific marker genes by comparing the expression patterns of different cell types, e.g., microglia vs. all other glia cells
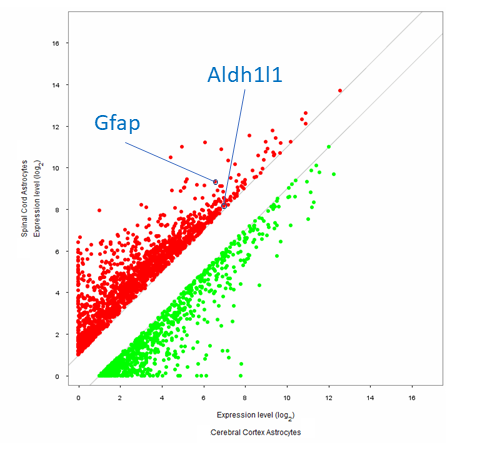
- Identify different subpopulations of a particular cell type by investigating gene expression changes between cells in different anatomical locations, e.g., brain astrocytes vs. spinal astrocytes (see image below)
- Characterize your genes of interest by comparing the gene expression pattern across multiple anatomical locations or grouping the genes by function with a gene set enrichment analysis
- Discover genes expressed together with your gene of interest by using a co-expression analysis
- Validate candidate genes from the analysis of this study in other single-cell or bulk-tissue studies