A recent pre-print study has identified several drugs that block SARS-CoV-2 replication based on their ability to counteract the virus-induced transcriptional landscape. In this study (https://doi.org/10.1101/2020.07.12.199687), the authors observed down-regulation of genes involved in cholesterol biosynthesis upon SARS-CoV-2 infection of lung adenocarcinoma cell line A549. The identified drugs (berbamine, amlodipine and others) were shown to counteract these transcriptional changes (i.e. up-regulate genes involved in cholesterol biosynthesis) and reduce viral load. The study data was curated by NEBION and is available in GENEVESTIGATOR® as HS-03527.
The Nebion compendium has been enriched with deeply curated in-vivo and in-vitro COVID-19/SARS-CoV-2 studies. Our unique tools enable scientists to explore transcriptomic signatures of the infection across patient samples, primary cells, organoids and cell line models across multiple studies in parallel. In the example below, we have selected a cluster of cholesterol biosynthesis genes up-regulated by these drugs and looked at regulation of these genes in the lungs of COVID patients and several in-vitro infection studies (including HS-03527 cited above). For some of the genes, we observed significant down-regulation upon infection in multiple studies, thus extending some of the findings to in-vivo infection and to other cell types such as intestinal organoids. However, we also noted up-regulation of some of these genes in COVID-19 patients with high viral load as compared to those with low viral loads, which is an unexpected finding warranting validation in further in vivo studies.
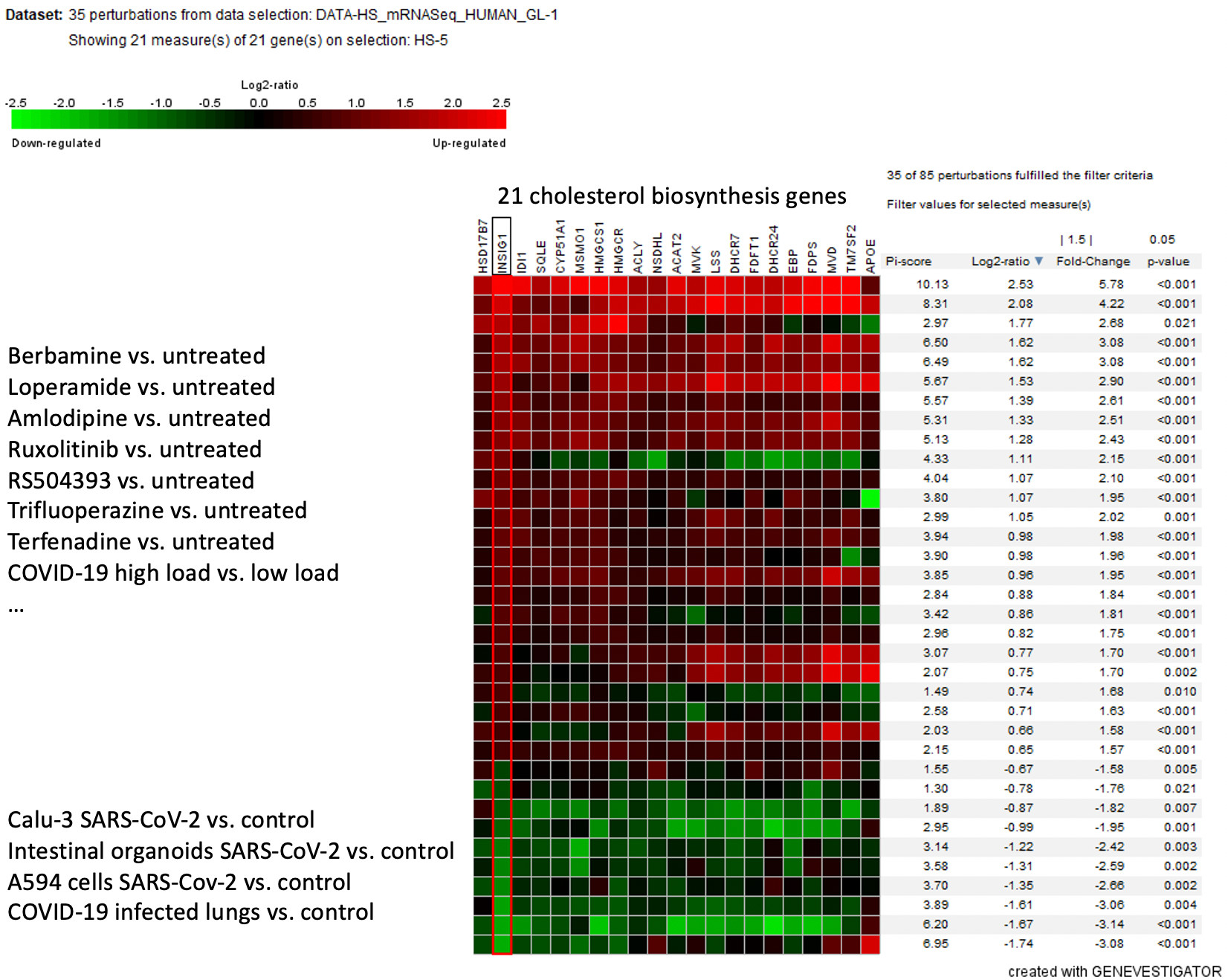
Figure: Regulation of cholesterol biosynthesis genes in SARS-CoV-2 and drug treatment studies. Data selection: human mRNA-seq coronavirus studies HS-03420, HS-03479, HS-3480, HS-3481, HS-3490, HS-03493, HS-03527, HS-03568. Condition search tool – perturbations. Filter: conditions affecting expression of INSIG1 (Fold change > 1.5, P-value < 0.05).